What is ORFEX?
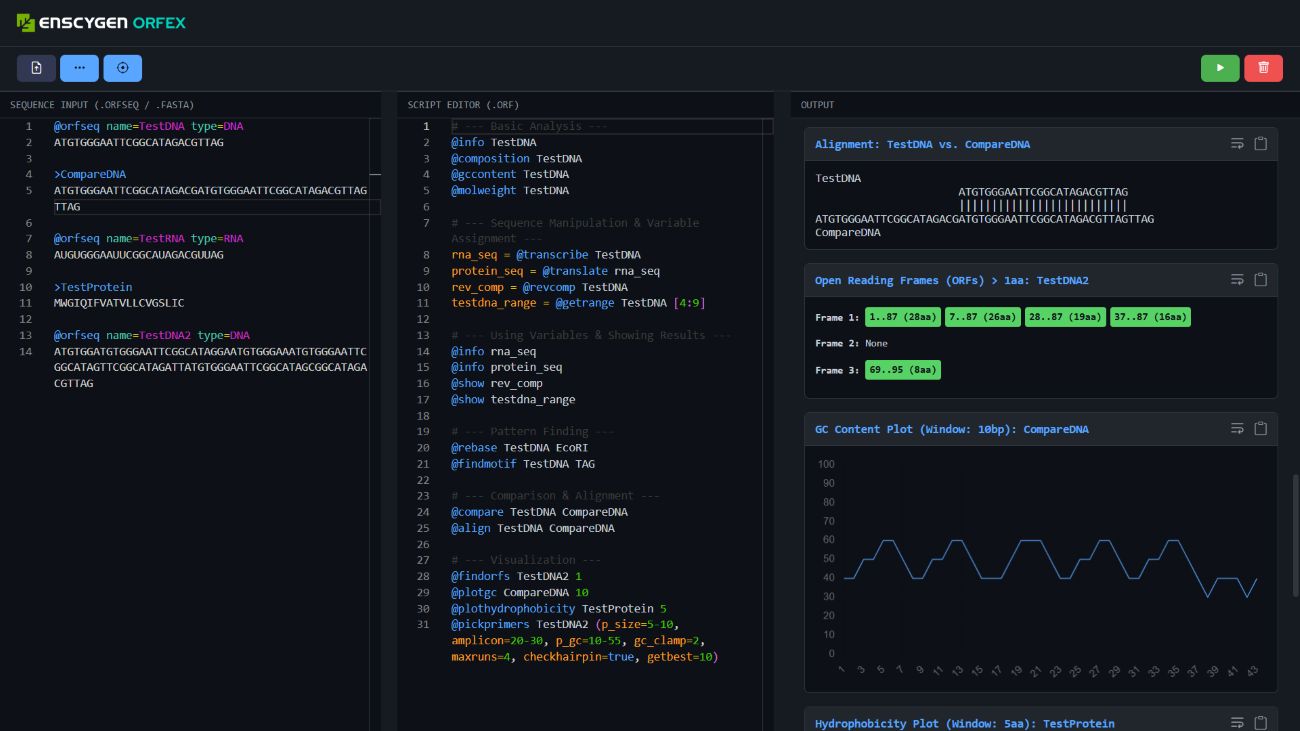
ORFEX is a novel programming language tailored for bioinformatics tasks involving DNA, RNA, and protein sequences. It is designed as both a scripting environment and a sequence notation system, making it ideal for educational, research, and computational biology applications.
Why ORFEX?
Unlike traditional tools that separate data from logic, ORFEX unifies biological sequences and commands in a code-driven format that is intuitive and editable. It's like VS Code meets a genome editor – interactive, scriptable, and visual.
Key File Types
- .orfex — Main script file (commands and logic)
- .orfseq — Sequence storage (DNA/RNA/Protein)
- .fasta — (Optional) Input support for sequence parsing
.orfseq Syntax
@orfseq name=Strand1 type=DNA
ATGGGCGTTTGA
@orfseq name=Strand2 type=RNA
AUGGCCUUUAA
@orfseq name=ProteinX type=Protein
MKWVTFISLLFLFSSAYS
Sequence Definition Commands
| Command Syntax | Description |
|---|---|
@orfseq name=MyGene type=DNA |
Defines a new DNA sequence with the name 'MyGene'. Type can be DNA,
RNA, or Protein.
|
>MyGene |
Defines a new sequence in FASTA format. Assumes type is DNA. |
Analysis & Manipulation Commands
| Command | Description |
|---|---|
@info MyGene |
Shows basic information about the sequence (e.g., length, type) in a table. |
@composition MyGene |
Generates a table showing the count and percentage of each base or amino acid. |
@gccontent MyGene |
Calculates the Guanine-Cytosine (GC) content percentage of a DNA sequence. |
@molweight MyGene |
Calculates the molecular weight (g/mol) of a DNA, RNA, or protein sequence. |
@transcribe MyGene |
Converts a DNA sequence into its corresponding RNA sequence by replacing Thymine (T) with Uracil (U). |
@translate MyGene |
Translates a DNA or RNA sequence into its corresponding amino acid sequence. |
@revcomp MyGene |
Generates the reverse complement of a DNA strand. |
@findmotif MyGene <Sequence> |
Finds all occurrences of a custom DNA sequence (motif). Example:
@findmotif MyGene GCCTA.
|
@rebase MyGene <EnzymeName> |
Finds all recognition sites for a given named restriction enzyme. Example enzymes:
EcoRI, BamHI.
|
Visualization Commands
| Command | Description |
|---|---|
@findorfs MyGene [minLength] |
Finds and displays all Open Reading Frames (ORFs) in a DNA/RNA sequence. An optional minimum length (in amino acids) can be provided. Default is 50. |
@plotgc MyGene [windowSize] |
Creates a sliding-window plot of the GC content across a DNA sequence. An optional window size can be provided. Default is 50. |
@plothydrophobicity ProteinX [windowSize] |
Generates a sliding-window plot of hydrophobicity for a protein sequence. Default window is 7. |
Use Cases
- Interactive gene/protein exploration
- Introductory bioinformatics coding practice
- Educational simulation tools for biology labs
- SSR detection, ORF mapping, synthetic strand design
Applications
ORFEX can be embedded in web-based tools, VS Code extensions, or used as a standalone educational interface. It is lightweight, interpretable via JavaScript or Python, and extensible with custom biological functions.